- 翰林提供学术活动、国际课程、科研项目一站式留学背景提升服务!
- 400 888 0080
IB DP Biology: SL复习笔记3.1.10 Skills: Using Databases
Use of Databases to Identify Gene Loci
Use of databases to identify the locus of a human gene and its polypeptide product
- Following the sequencing of the whole human genome, we now know the exact locus (position) of every gene across the 23 pairs of chromosomes
- Online databases have been built that are able to locate any known gene or allele
- Anyone can access these loci
- One example is the European Molecular Biology Laboratory database (EMBL)
- Examples of genes that can be located are
- The CFTR protein, critical to cystic fibrosis, on chromosome 7
- HBB, a faulty allele of which is the cause of sickle-cell anaemia, on chromosome 11
- If we know the locus of a particular gene, medicine can establish the location of a faulty allele, which is often recessive
- A faulty allele can be cut out of the chromosome by genetic engineering using recombinant DNA technology
- Replacing a faulty allele could lead to genetic therapy
- Location databases of cancer-related genes are often vital information to researchers, doctors and patients involved in cancer genetics
Use of Databases: Comparing Base Sequences
Use of a database to determine differences in the base sequence of a gene in two species
- The Genbank® database is another that can be used to search for DNA base sequences
- Uses a computer data analysis technique called BLAST (Basic Local Alignment Search Tool) to spot and 'line up' similar base sequences
- A protein common to all organisms is cytochrome C
- This makes its gene sequence a good one to compare between organisms
- The sequence is available for many different organisms across all three domains
- This gives important information about evolutionary relationships between organisms
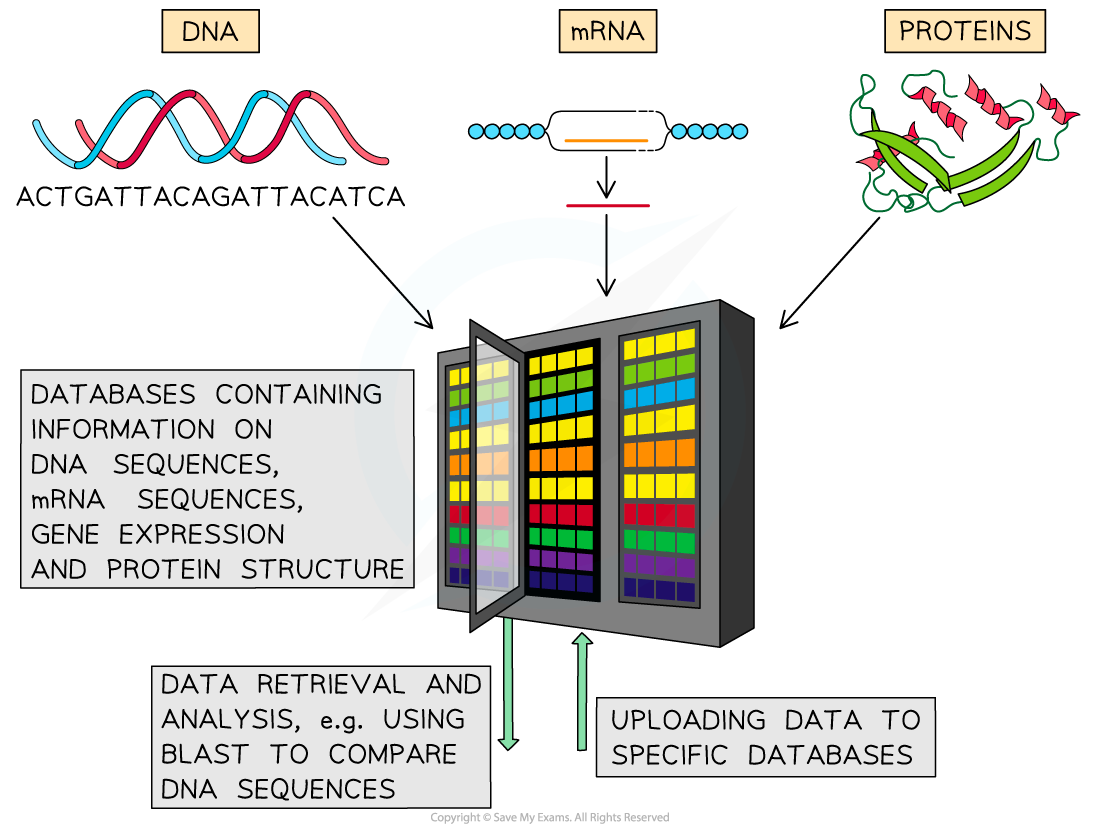
The use of databases to compare base sequences (and protein sequences) between species
转载自savemyexams
站内搜索
竞赛真题免费下载(点击下载)
在线登记
最新发布
© 2024. All Rights Reserved. 沪ICP备2023009024号-1





